Unlike extensively-studied carbon (C) flux map of plant metabolism, little is known how assimilated N flows through the metabolic network, namely N flux map (NFM). As N metabolism can strictly constraint C metabolism in plants, the integration of the NFM into the plant metabolic models will be crucial to understand and improve plant metabolism and productivity especially under limited N input. The goals of this project are to construct plant NFMs from plant genomes and to determine both biochemical and system-level functionality of plant N metabolic network.
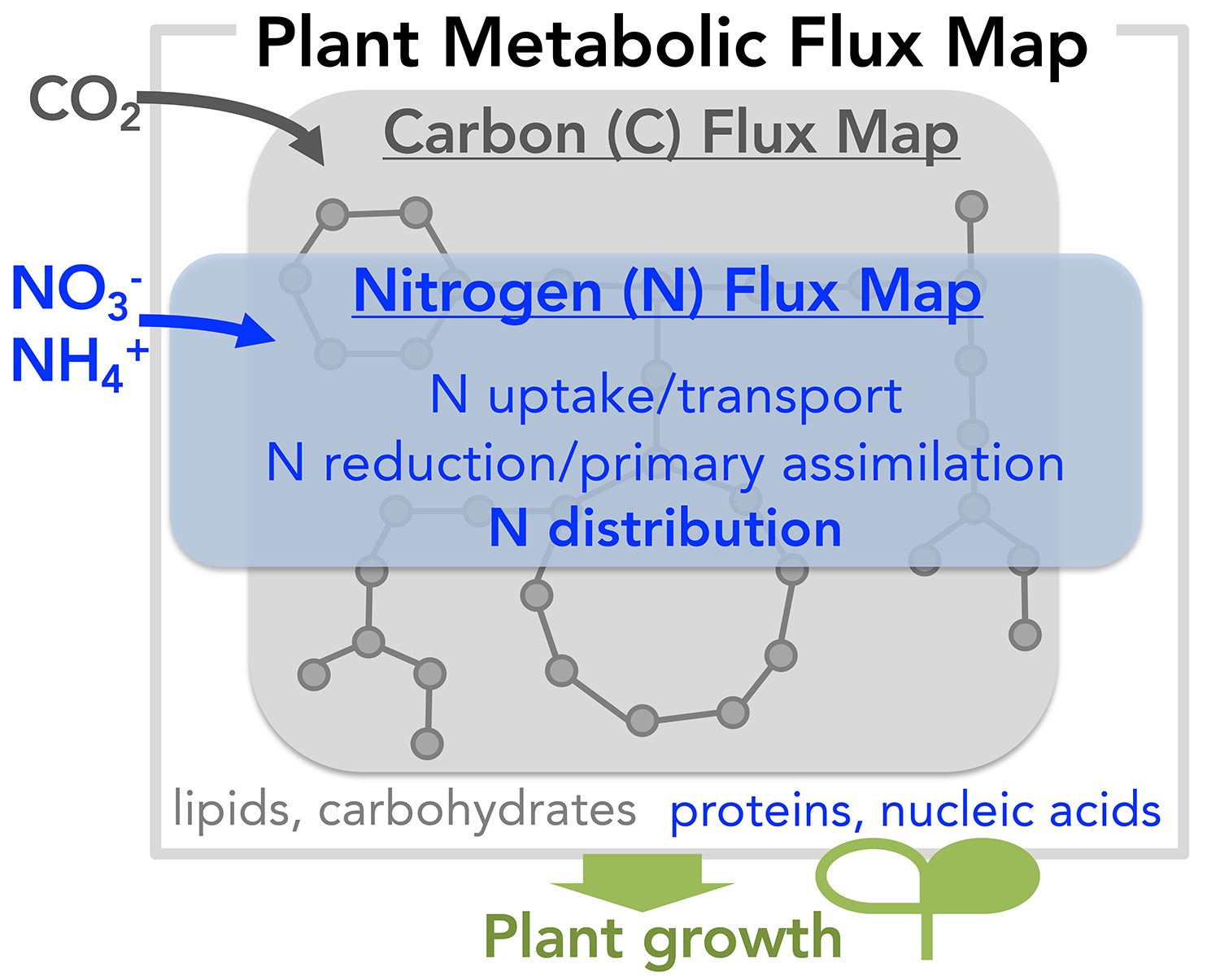
While the early steps of N metabolism, i.e. inorganic N uptake and assimilation, have been extensively studied, much less is known how assimilated N is distributed throughout the plant metabolic network. The core of NFM is composed of different branches of amino acid metabolism interconnected by aminotransferases (ATs). AT enzymes play pivotal roles in distributing reduced N for synthesis of various organonitrogen compounds, such as amino acids and their downstream products including proteins, nucleic acids, and alkaloids. Genes and enzymes responsible for specific AT reactions have been identified. However, an AT enzyme can potentially catalyze 380 different transamination reactions simply considering 20 proteogenic amino acids and corresponding keto acids as potential substrates. Yet, the multi-substrate specificities of ATs remain largely uncharacterized due to their poor sequence-function relationships and tedious aminotransferase activity assays that allow testing of only “two substrates at a time”. As a result, we do not know the true functionality of AT enzymes and how ATs transfer N across the plant N metabolic network, i.e. NFM.
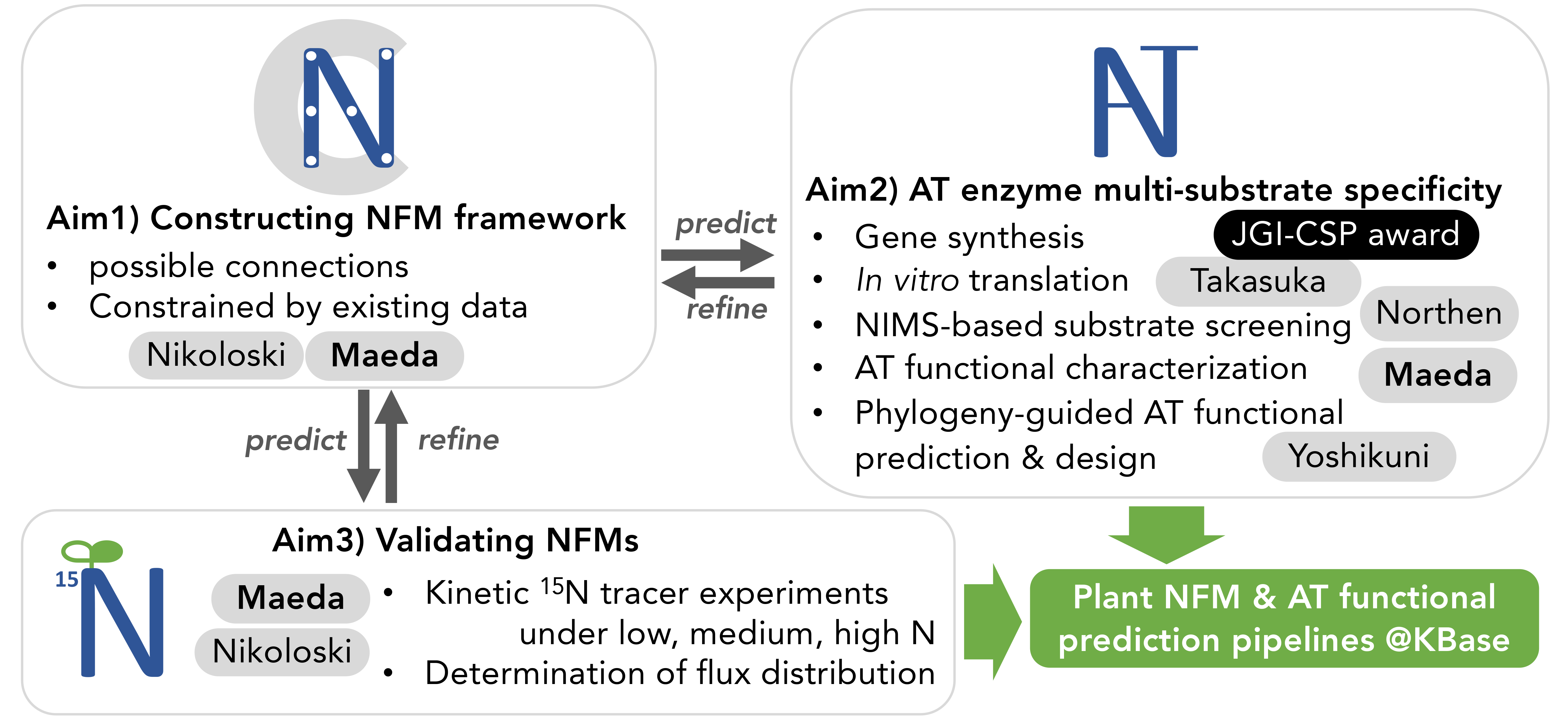
To address this grand challenge, our project makes use of rapidly growing numbers of plant genomes, high-throughput functional characterization platforms, and computational modeling to deduce both biochemical and systems level functionality of ATs and NFMs. Aim 1 will construct a biochemically feasible NFM from the plant genomes using computational modeling. Aim 2 will experimentally determine biochemical functions of AT enzymes through high-throughput gene/protein synthesis and enzyme assay platforms, followed by computational protein modeling to predict AT functions from other species. Aim 3 will validate the system-level functionality of ATs and will determine N flux distribution under different N availability through kinetic stable isotope precursor feeding. The resulting NFMs will serve as a novel framework to i) elucidate how N flows through the plant metabolic network in a quantitative manner, ii) simulate how plant metabolism responds to different N availability at a systems level, and iii) identify potential targets for improving N use efficiency. We will also establish open source public databases and pipelines in DOE Systems Biology Knowledgebase (KBase) for other researchers to be able to predict AT functions and construct NFMs from any given plant genomes.